RNA-seq Analysis Reveals Mode of Splicing Inefficiency: Coordinated Intron Retention
Physical and Biological Sciences
Department of Molecular, Cell and Developmental Biology
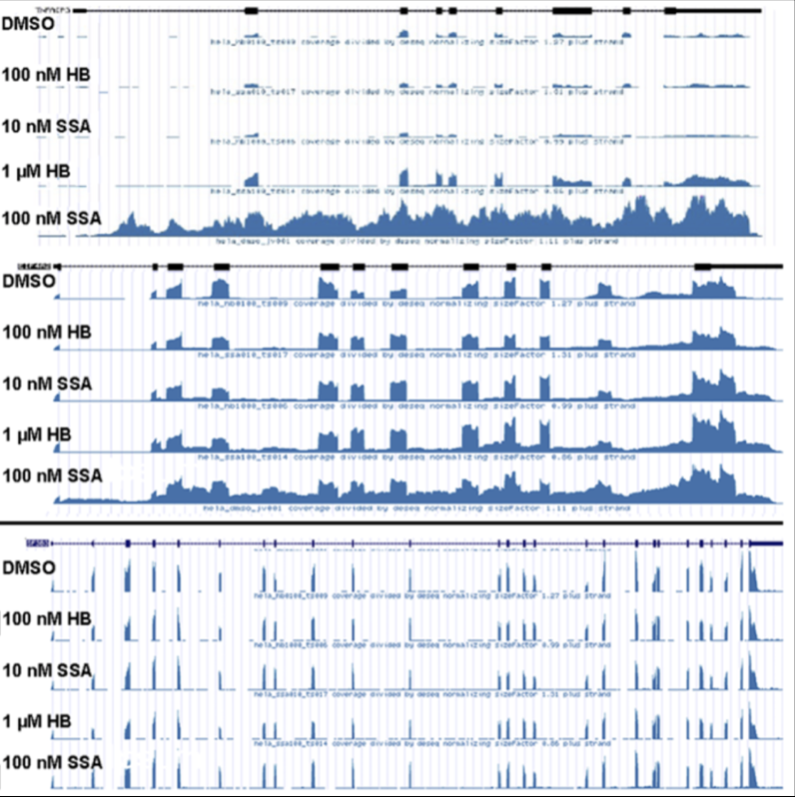
Recently, an anti-tumor drug targeting a key component of the spliceosome has been identified: spliceostatin A (SSA). However, the mechanism of splicing interruption and regulation on gene expression due to SSA has not been characterized. To understand how SSA can inhibit cancer cell growth, RNA-seq data provides a rich resource for the detection of alternative splicing events. In this approach, an algorithm was built to test whether splicing inhibitors lead to a new mode of splicing inefficiency characterized by coordinated intron retention (IR) events across an entire transcript. Transcriptome wide analysis was performed to gain a deeper insight into full detection of IR events among all transcripts (rather than known IR events). Indeed, cells treated with SSA expressed a greater number of transcripts showing splicing inefficiency in contrast to non-treated cells. Unexpectedly, SSA RNA-seq captured a binary distinction of IR such that all introns of a transcript are retained/coordinated or none are affected. A subset of gene transcripts involved in the innate immune response were observed to contain upregulated coordinated IR events. As a whole, results from this study offers a starting point to help guide chemotherapy development by offering insight into the effects on splicing.