Pan-Cancer Analysis of Driver Mutations and Modules in Cancer Genomes
Physical and Biological Sciences
Genomics Institute RMI program
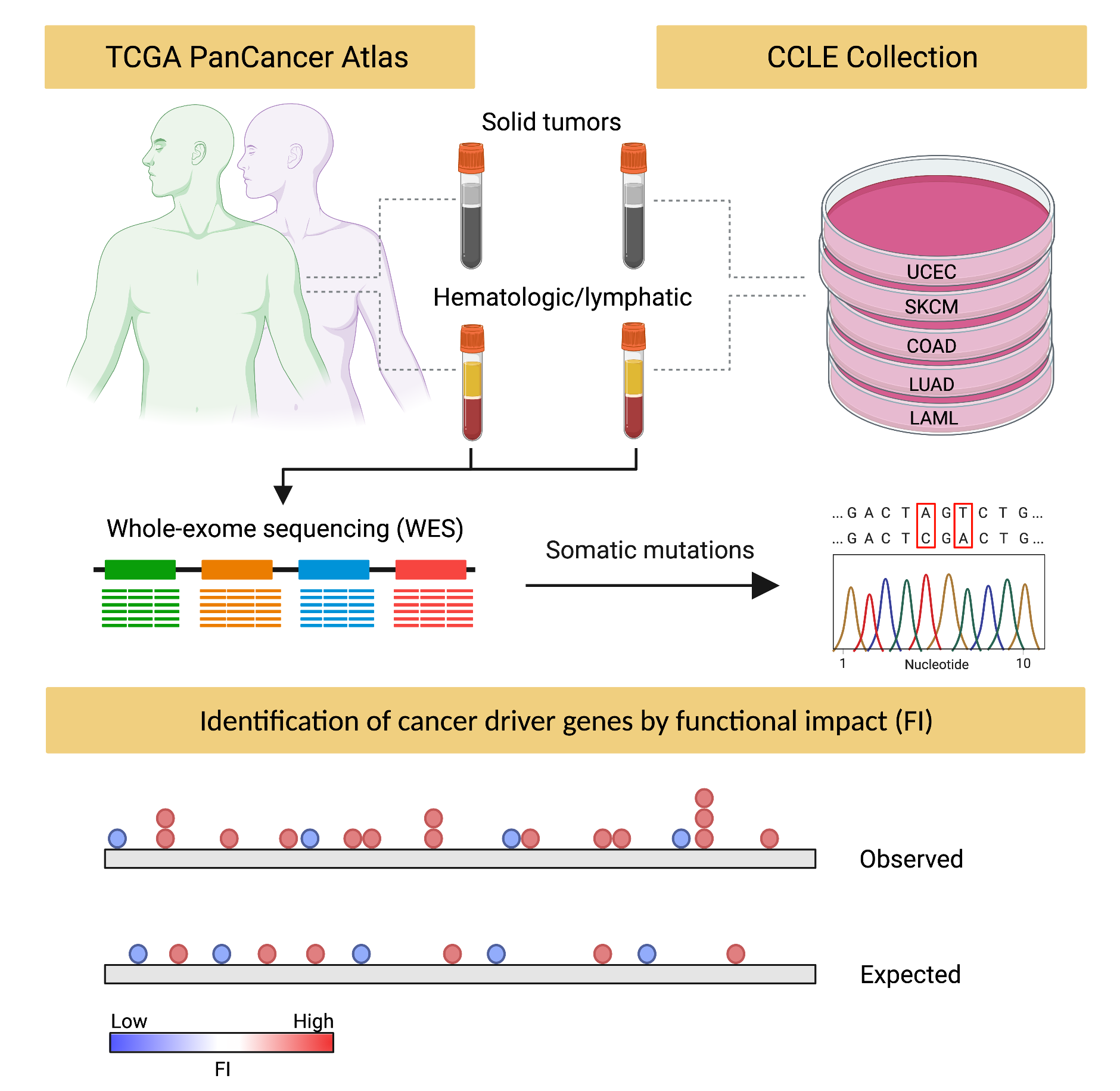
Cancer driver mutations and modules are central to understanding tumorigenesis, tumor malignancy, recurrence, and drug resistance. However, driver mutations and modules across tumor types are not completely characterized nor understood due to limitations of tumor samples and the complexity of biological pathways. Here, we present a comprehensive analysis of driver mutations and modules in cancer genomes across 49 cancer types from The Cancer Genome Atlas (TCGA) Pan-Cancer studies, and Cancer Cell Line Encyclopedia (CCLE). We used whole- exome somatic mutation variants from TCGA and CCLE to characterize a landscape of driver alterations across cancers in protein coding genomic regions. We identified well-known and studied driver mutations and revealed previously unannotated driver genes. TP53 was the most significant and mutated driver across tumors, followed by KRAS, PTEN, RB1, CDKN2A, NRAS, CASP8, and STK11. DSPP and PTPRQ were the only unannotated driver gene in the Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census (CGC), which were highly significant in both TCGA and CCLE pan-cancer analysis. We also observed highly significant and potentially unannotated driver genes such as ELAVL1, FHL2, GNB2L1, DNAH8 and DNAH9. Many of these possible unannotated drivers, such as DSPP, have been previously suggested as potential cancer driver genes using experimental approaches, however, they have not been previously characterized as drivers across all cancer types. This study provides a model and comprehensive landscape of driver genes and mutations across cancer type genomes which might serve as an asset for future research and clinical endeavors.