Ancestry HMM with Gene Conversion
Engineering
Biomolecular Engineering
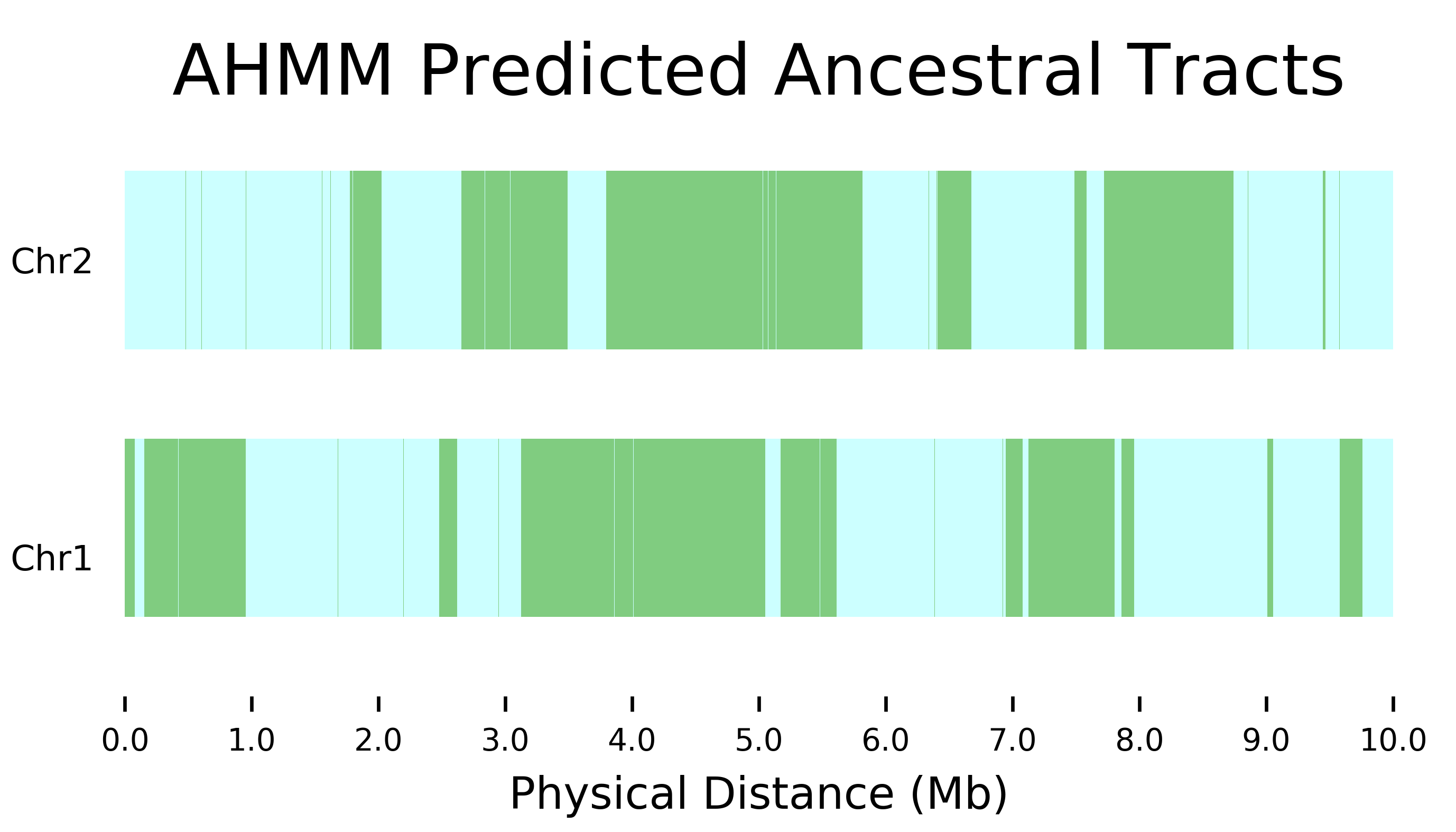
Admixture, the result of interbreeding between genetically distinct populations, can give an early glimpse into reproductive incompatibility and speciation processes (Corbett-Detig et al., 2017; Medina et al., 2018). When recombination introduces breaks in ancestry along a chromosome, the resulting ancestry tracts yield valuable information about the admixture process. For example, in more ancient admixture, there is more time for recombination, and the resulting tracts should be shorter. In particular these signatures of recombination can be inferred and studied using probabilistic models, but this picture is incomplete because double strand breaks during meiotic recombination can induce gene conversions as well as meiotic crossovers (Korunes & Noor, 2016). Gene conversion is a strong evolutionary force, especially over short genetic distances, that is thought to have shaped Eukaryotic genomes and our meiotic machinery (Burt & Trivers, 2008). Hence, we have updated the inference model of Ancestry HMM to include the gene conversion process in a comprehensive recombination model. For diploid individuals sampled from our simulated admixed population, over 85% of individual gene conversion segments are detected down to single variant resolution. We demonstrate that the gene conversion process is modelled well by our HMM across various simulated data parameters using both curated genotype and read pile-up data. Finally, we employ a Nelder-Mead parameter optimization algorithm for learning the unknown parameters that generated the data. This will enable researchers to identify and study gene conversion across species with the genetic map also serving as an effective scaffold in de novo genome assembly applications.